

本年度在学校的支持下,清华大学合成与系统生物学中心取得了成绩,列举如下:
1、使用 CRISPRi技术实现了对细胞形态的控制,对提高生物技术的产量将产生很大的作用:(陈国强)
图1:用CRISPRi技术调控细胞X和Y轴的大小,从而实现对细胞大小的控制。对生物技术的产业化将有大的推动。
2、构建诱导型人工超强启动子,原位替换野生枯草芽孢杆菌基因组上的表面活性素合成酶启动子,实现了表面活性素产量提高17.7倍。(于慧敏)
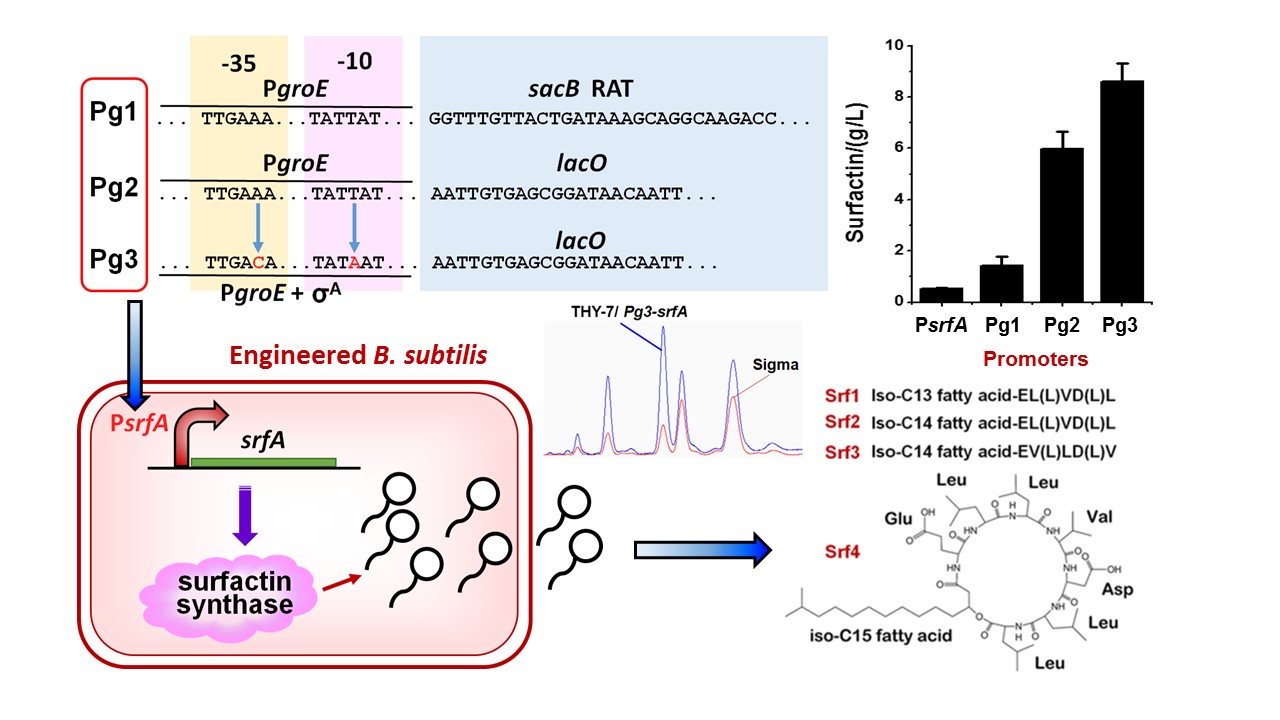
图2: 设计构建诱导型人工超强启动子Pg3实现重组枯草芽孢杆菌高产脂肽类表面活性素。
3. 建立了多组分蛋白质复合物共表达系统,极大的缩短了蛋白质复合物的构建和纯化所需的时间:(戴俊彪)
图3:利用EcoExpress进行蛋白复合物克隆、表达和纯化的过程。对复合物晶体及电镜结构起到促进作用。
4. 高通量微生物进化仪的研制取得新突破(邢新会)
在ARTP诱变育种模块的基础上,成功研制“微液滴恒化培养”模块,并进入商业化样机开发阶段,这将是全球首个基于微流控技术的微生物恒化培养设备。本模块可以平行实现200个微生物液滴的恒化培养和生长参数采集,可以高通量的进行微生物适应性进化、抗生素耐受性演变、微生物生长特性采集等多方面研究。

图4:高通量微生物进化仪
5. 甲醇唯一碳源微生物扭脱甲基杆菌AM1细胞工厂改造新进展(张翀)
开发了基于生物传感器辅助的全局转录因子工程新方法,借助甲羟戊酸传感器,超高通量筛选和改造全局转录因子QscR,“四两拨千斤”的提高目的产物甲羟戊酸产量,解决了甲基营养菌中循环代谢途径节点Ac-CoA代谢流再分配的难题。
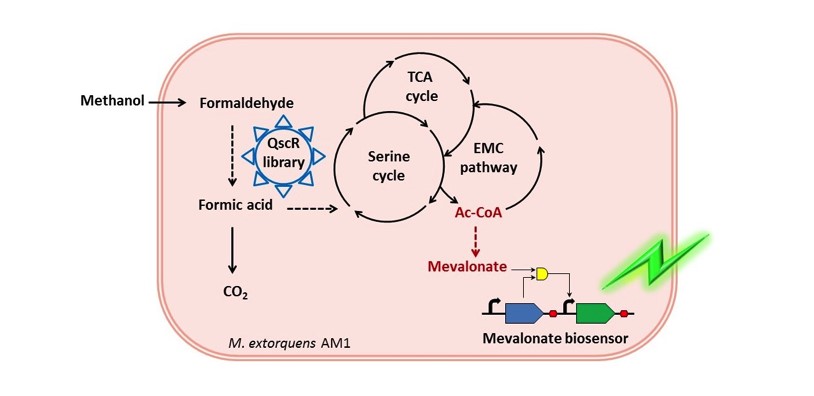
图5:以甲醇为唯一碳源的细胞工厂示意图
6.以活性炭为载体高效固定化酶新方法(刘铮)
活性炭广泛应用于各类化工过程,如果能够将酶固定中活性炭上,将会极大地拓展酶催化中化工过程中的应用。以活性炭来固定化酶的研究最早见诸于上世纪初,但始终受制于如下两个问题:一是如何降低酶中活性炭表面的失活;二是如何抑制酶从活性炭孔内泄露出来。本研究采用刀豆蛋白(Concanavalin A, ConA)对含有糖残基的酶的分子识别和凝集作用将酶稳定地吸附在活性炭孔内。采用辣根过氧化物酶(HRP)和碳酸酐酶(CA)的实验结果表明,Con A可以有效地降低酶在活性炭孔表面的失活;且形成的酶聚集体可以稳定地包埋在活性炭孔内而不泄露;重复使用批次可以到25次以上;固定化酶具有更宽泛的操作pH和温度范围;而活性炭宽泛的吸附功能也使得这类固定化酶对于低浓度底物转化具有优势。
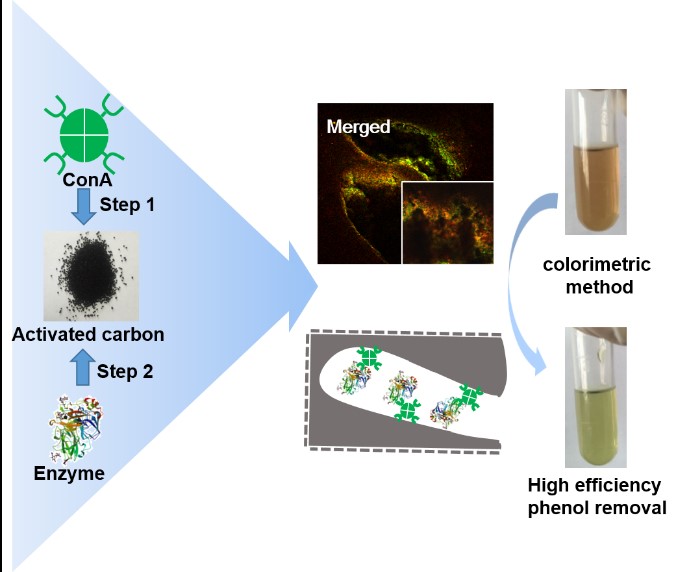
图6. 以活性炭为载体高效固定化酶新方法
7. 高活性、可回收使用的磁性双酶纳米催化剂(刘铮)
双酶或者多酶催化剂的构建对于拓展工业酶催化具有重要的基础意义,为提高其催化活性需要拉近酶分子间距以强化底物和中间产物传递;为便于工业使用需要解决酶催化剂回收问题。磁性纳米颗粒是一个很好的载体,但酶分子在其表面会失活。本研究提出首先将ConA交联到磁性纳米颗粒表面,再吸附上酶分子并进行交联的方法来制备磁性双酶纳米催化剂。采用葡萄糖氧化酶(GOx)和辣根过氧化物酶(HRP);以及 GOx和氧化酶 (Cat)构成的双酶体系证明了上述方法的可行性;酶分子间距离可缩短至6.4nm;显著地提高了表观催化活性。而对于GOx-Cat双酶体系,还可出现有害中间产物及时消解和底物循环效应,由此提升酶催化总体效率;利用磁场可以方便的实现酶催化剂的回收。上述成果已经中本年度的美国化学工程师学会年会上口头报告(Paper 650g),目前正在 ACS Catalysis 审稿中。
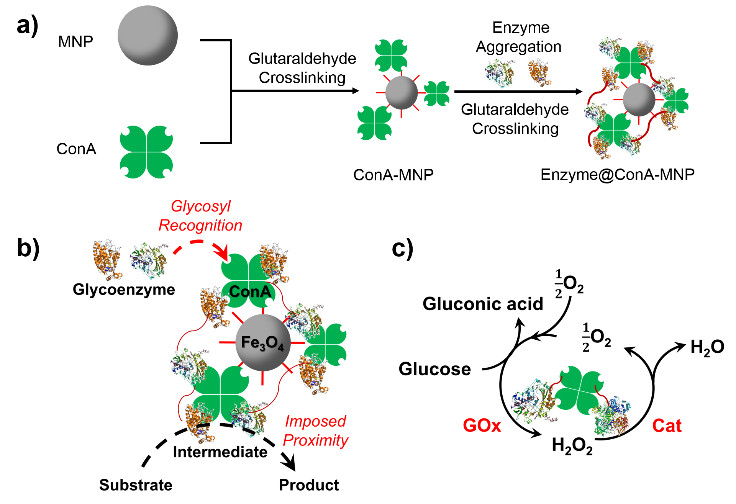
图7. 磁性双酶纳米催化剂
8. 实现年产2万吨1,3-丙二醇的规模化工业生产,产品顺利用于PTT的聚合及下游纺丝(刘德华):

图8. 1,3-丙二醇的规模化工业生产
9. 对影响衰老的两个通路进行了研究。发现NAD的降低可以影响上皮间质细胞转换过程而蛋白合成的下调提高细胞耐受压力的功能(邓海腾)。
10. CRISPR/Cas9系统的精细调控(谢震)
CRISPR/Cas9系统是在细菌中起到抵御外源DNA入侵的作用,通过人工设计靶向特定序列的guide RNA(靶向RNA),人们可以利用Cas9蛋白进行基因组的特定位点的切割。不仅如此,基于失去核酸酶活性的dCas9,通过与不同效应蛋白结构域偶联,可以实现特定基因位点基因的激活和抑制。然而CRISPR/Cas9系统目前还缺乏更为精细的控制,从而实现在特定细胞中发挥作用。谢震课题组通过利用内含肽将Cas9进行合理的拆分,从而可以实现三输入“与门”逻辑。同时,利用蛋白质结构域可控交换的策略,实现了在不同细胞中对一个基因和两个基因不同的控制,这为进一步实现Cas9在医疗上的安全应用奠定了基础。该成果已在Nature Communications杂志发表, 并已申请一项中国专利和一项美国专利。
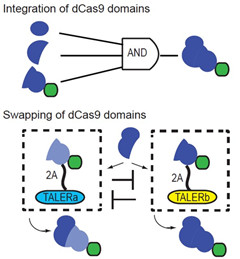
图10. 拆分CRISPR/Cas9系统实现三输入“与门”逻辑
11. 竞争性调控:没有物理相互作用的生物两类分子由于共享有限资源而相互影响和调控
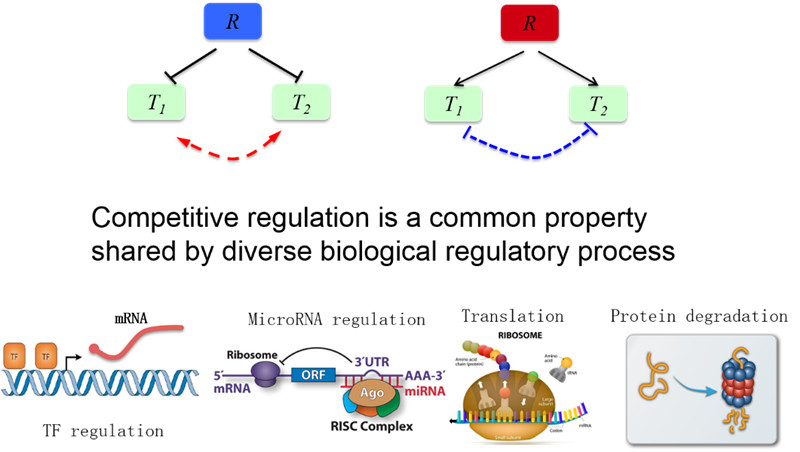
图11. 竞争性调控示意图(汪小我)
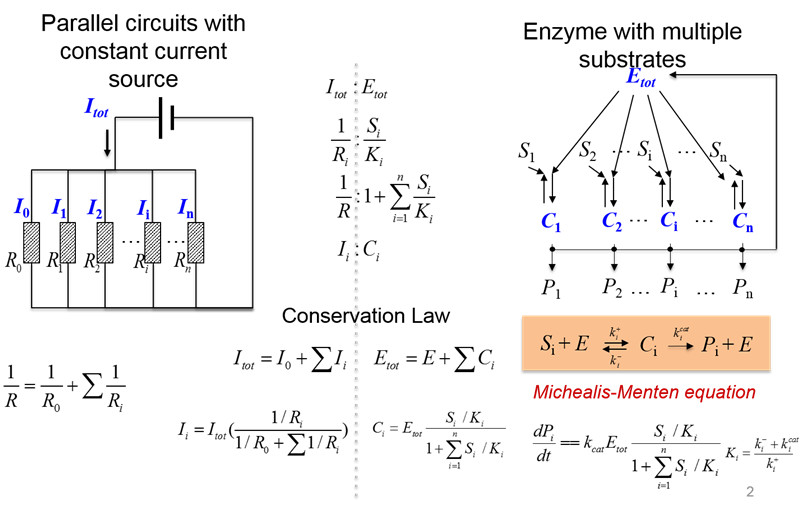
图12. 竞争性调控与平行电路中的电流分配类似(汪小我)
CSSB论文发表:
1. Li, T.T., Li, T., Ji, W.Y., Wang, Q.Y., Zhang, H.Q., Chen, G.Q., Lou, C.B., Ouyang, Q., 2016. Engineering of core promoter regions enables the construction of constitutive and inducible promoters in Halomonas sp. Biotechnol. J. 11, 219-227.
2. Jiang, X.R., Chen, G.Q., 2016. Morphology engineering of bacteria for bio-production. Biotechnol. Adv. 34, 435–440.
3. Chen, G.Q., 2016. Omics meets metabolic pathway engineering. Cell Syst. 2, 335-346.
4. Wu, H., Fan, Z.Y., Jiang, X.R., Chen, J.C., Chen, G.Q., 2016. Enhanced production of Polyhydroxybutyrate by multiple dividing E. coli. Microb. Cell Fact. 15, 128.
5. Ma, Y.M., Yao, H., Wei, D.X., Wu, L.P., Chen, G.Q., 2016. Synthesis, characterization and application of comb-like Polyhydroxyalkanoate-graft-Poly(N-isopropylacrylamide). Biomacromolecules 17, 2680-2690.
6. Hajnal, I., Chen, X.B., Chen, G.Q., 2016. A novel cell autolysis system for cost-competitive downstream processing. Appl. Microbio. Biotechnol. 100, 9103-9110.
7. Elhadi, D., Lv, L., Jiang, X.R., Wu, H., Chen, G.Q., 2016. CRISPRi engineering E. coli for morphology diversification. Metab. Eng. 38, 358-369.
8. Wu, H., Chen, J.C., Chen, G.Q., 2016. Engineering the growth pattern and cell morphology for enhanced PHB production by Escherichia coli. Appl. Microbiol. Biotechnol. 100, 9907-9916.
9. Zhang, S.Q., Fang, L., Li, Z.J., Wu, Q., Guo, Y.Y., Chen, G.Q., 2016. Enhanced production of Poly-3-hydroxybutyrate by recombinant Escherichia coli containing NAD kinase and phbCAB operon. Science China Chemistry 59, 1390-1396.
10. Kwan, T.D., Markides, H., Wang, W.B., Yu. T., Chen, GQ., Liu, W., Haj, A.J., 2016. The application of nanomagnetic approaches for targeting adipose derived stem cells for use in tendon repair. J. Nanosci. Nanotechnol. 16, 8989-8999.
11. Chen, G.Q., Jiang, X.R., Guo, Y.Y, 2016. Synthetic biology of microbes synthesizing Polyhydroxyalkanoates (PHA). Synthetic and Systems Biotechnology 1, 236-242.
12. Yao, H., Wei, D.X., Ma, Y.M., Cai, L.W., Tao, L., Liu, L., Wu, L.P., Chen, G.Q., 2016. Comb-like temperature-responsive Polyhydroxyalkanoate-graft-Poly(2-Dimethylamino- ethylmethacrylate) for controllable protein adsorption. Polymer Chemistry 7, 5957-5965.
13. Li, T., Ye, J.W., Shen, R., Zong, Y.Q., Zhao, X.J., Luo, C.B., Chen, G.Q., 2016. Semi-rational approach combining one-step library construction and high throughput screening for ultrahigh production of poly(3-hydroxybutyrate) by Escherichia coli. ACS Synth. Biol. 5, 1308-1317.
14. Li, T., Guo, Y.Y., Qiao, G.Q., Wu, Q., Chen, G.Q., 2016. Microbial synthesis of 5-aminolevulinic acid (ALA) and its co-production with polyhydroxybutyrate (PHB). ACS Synth. Biol. 5, 1264-1274.
15. Lan, L.H., Zhao, H., Chen, J.C., Chen, G.Q., 2016. Halomonas as a low-cost production host for production of bio-surfactant protein PhaP. Biotechnol. J.. 11, 1595-1604.
16. Ren, Y.L., Meng, D.C., Chen, J.C., Wu, Q., Chen, G.Q., 2016. Microbial synthesis of a novel terpolyester P(LA-co-3HB-co-3HP) from low cost substrates. Microbial. Biotechnol. DOI:10.1111/1751-7915.12453
17. Zhao, H., Zhang H.Q., Li, T., Lou, C.B., Ouyang, Q., Chen, G.Q., 2016. Novel T7-like expression systems used for Halomonas. Metab. Eng. S1096-7176, 30136-30127.
18. Zhao, H.Y., Wang, X.Q., Chen, G.Q., 2016. Structural Insights on PHA Binding Protein PhaP from Aeromonas hydrophila. Sci. Rep. doi: 10.1038/srep39424 (2016)
19. Zhang, Y.A., Ma, X., Sathe, A., Fujimoto, J., Wistuba, II., Lam, S., Yatabe, Y., Wang, Y.W., Stastny, V., Gao, B., Larsen, J.E., Girard, L., Liu, X., Song, K., Behrens, C., Kalhor, N., Xie, Y., Zhang, M.Q., Minna, J.D., Gazdar, A.F., 2016. Validation of SCT Methylation as a Hallmark Biomarker for Lung Cancers. J. Thorac. Oncol. 1,346-60.
20. Khan A., Zhang, X., 2016. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 44, D164-71.
21. Feng, H., Li, T., Zhang, X., 2016. Characterization of kinase gene expression and splicing profile in prostate cancer with RNA-Seq data. http://biorxiv.org. doi: http://dx.doi.org/10.1101/061085.
22. Zhang, X., Liu, S., Cui, H., Chen, T., 2016. Reading the underlying information from massive metagenomic sequencing data. Proc. IEEE. 99,1-15.
23. Hong, F., Xue,Y., Zhang, X., 2016. An overview of major metagenomic studies on human microbiomes in health and disease. Quantitative Biology. 4, 192–206.
24. Mangericao T.C., Peng, Z., Zhang, X., 2016. Computational prediction of CRISPR cassettes in gut metagenome samples from Chinese type-2 diabetic patients and healthy controls. BMC Syst. Biol. Suppl1, 5.
25. Wang, B., Ji, T., Zhou, X., Wang, J., Wang, X., Wang, J., Zhu, D., Zhang, X., Sham, P.C., Zhang, X., Ma, X., Jiang, Y., 2016. CNV analysis in Chinese children of mental retardation highlights a sex differentiation in parental contribution to de novo and inherited mutational burdens. Sci. Rep. 6, 25954.
26. Chen, H., Zhou, X., Wang, J., Wang, X., Liu, L., Wu, S., Li, T., Chen, S., Yang, J., Sham, P.C., Zhu, G., Zhang, X., Wang, B., 2016. Exome Sequencing and Gene Prioritization Correct Misdiagnosis in a Chinese Kindred with Familial Amyloid Polyneuropathy. Sci. Rep. 6, 26362.
27. Jiang, B., Ye, C., Liu, J.S., 2016. Bayesian Nonparametric Tests via Sliced Inverse Modeling. Bayesian Analysis. TBA, 1–24.
28. Zhang, L., Liu, X., Zhang, X., Chen, R., 2016. Identification of important long non-coding RNAs and highly recurrent aberrantal ternative splicing events in hepatocellular carcinoma through integrative analysis of multiple RNA-Seq datasets. Mol. Genet. Genomics. 291,1035-51.
29. Jiao, S., Li, X., Yu, H., Yang, H., Li, X., Shen, Z., 2016. In situ enhancement of surfactin biosynthesis in Bacillus subtilis using novel artificial inducible promoters. Biotechnol Bioeng. doi: 10.1002/bit.26197. [Epub ahead of print]
30. Sun, J., Yu, H., Chen, J., Luo, H., Shen, Z., 2016. Ammonium acrylate biomanufacturing by an engineered Rhodococcus ruber with nitrilase overexpression and double-knockout of nitrile hydratase and amidase. J. Ind. Microbiol. Biotechnol. 43,1631-1639.
31. He, X., Dai, J., and Wu, Q., 2016. Identification of sporopollenin as the outer layer of cell wall in Microalga Chlorella protothecoides. Front Microbiol. 7, 1047. (*Co-corresponding author)
32. Qin, Y., Tan, C., Lin, J., Qin, Q., He, J., Wu, Q., Cai. Y., Chen, Z.,* Dai, J.,* 2016. EcoExpress—highly efficient construction and expression of multicomponent protein complexes in Escherichia coli. ACS Synth. Biol. [Epub ahead of print] (*Co-corresponding author)
33. Martella, A.,* Pollard, S.M., Dai. J., Cai, Y.,* 2016. Mammalian synthetic biology: time for Big MACs. ACS Synth. Biol. 5, 1040-1049.
34. Yan, Y.S., Ji, Y., Su, N., Mei, X., Wang, Y., Du, S.S., Zhu, W.M., Zhang, C., Lu, Y., Xing, X.H., 2016. Non-anticoagulant effects of low molecular weight heparins in inflammatory disorders: a review. Carbohydrate Polymers. In press. DOI:10.1016/j.carbpol.2016.12.037.
35. Wang, C.H., Zhang, C., Xing, X.H., 2016. Xanthine dehydrogenase: An old enzyme with new knowledge and prospects. Bioengineered 7, 395-405.
36. Wang, C.H., Li, G., Zhang, C., Xing, X.H., 2016. Enhanced catalytic properties of novel (abg)2 heterohexameric Rhodobacter capsulatus xanthine dehydrogenase by separate expression of the redox domains in Escherichia coli. Biochemical Eng. J. In press.
37. Liang, W.F., Cui, L.Y., Cui, J.Y., Yu, K.W., Yang, S., Zhang, C.*, Xing, X.H. 2016 Biosensor-assisted transcriptional regulator engineering for Methylobacteriumextorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply. Metab. Eng. doi:10.1016/j.ymben.2016.11.010
38. Fang, M., Wang, T., Zhang, C.*, Bai, J., Zheng, X., Zhao, X., Xing, X.H., 2016. Intermediate-sensor assisted push-pull strategy and its application in heterologous deoxyviolacein production in Escherichia coli. Metab. Eng. 33, 41–51.
39. Xu, W.N., Yong, Y., Wang, Z.Y., Jiang, G.Q., Wu, J.Z., Liu, Z., 2016. Concanavalin a coated activated carbon for high performance enzymatic catalysis. ACS Sustainable Chem & Eng. DOI:10.1021/acssuschemeng.6b02705.
40. Chen, Z., Huang, J.H., Wu, Y., Wu, W., Zhang, Y., Liu, D.H., 2016. Metabolic engineering of Corynebacterium glutamicum for the production of 3-hydroxypropionic acid from glucose and xylose. Metab. Eng. DOI: 10.1016/j.ymben.2016.11.009.
41. Chen, Z., Liu, D.H., 2016. Toward glycerol biorefinery: metabolic engineering for the production of biofuels and chemicals from glycerol. Biotechnol. Biofuels 9, 205.
42. Chen, Z., Huang, J., Wu, Y., Liu, D.H., 2016. Metabolic engineering of Corynebacterium glutamicum for the de novo production of ethylene glycol from glucose. Metab. Eng. 33, 12-18.
43. Chen, H.M, Zhao, X.B., Liu, D.H., 2016. Relative significance of the negative impacts of hemicelluloses on enzymatic cellulose hydrolysis is dependent on lignin content: evidences from substrate structural features and protein adsorption ACS Sustainable Chemistry Eng. DOI: 10.1021/acssuschemeng.6b01540
44. Akimkulova, A., Zhou, Y., Zhao, X.B, Liu, D.H., 2016. Improving the enzymatic hydrolysis of dilute acid pretreated wheat straw by metal ion blocking of non-productive cellulase adsorption on lignin. Bioresour. Technol. 208, 110-116.
45. Wu, R.C., Zhao, X.B., Liu, D.H., 2016. Structural features of formiline pretreated sugarcane bagasse and their impacts on the enzymatic digestibility of cellulose. ACS Sustainable Chemistry Eng. 4, 1255–1261.
46. Qi, F., Zhao, X.B., Li, T., Ou, X.J., Du, W., Liu, D.H., 2016. Integrative transcriptomic and proteomic analysis of the mutant lignocellulosic hydrolyzate-tolerant Rhodosporidium toruloides. Engineering in Life Science DOI: 10.1002/elsc.201500143.
47. Liu, Z. L., Hu, Y. D., Wang, H. L., Liu, C. D., Wang, Q. T., Deng, H., 2016. Hydrogen peroxide mediated mitochondrial UNG1-PRDX3 interaction and UNG1 degradation. Free Radic Biol. Med. 99, 54-62.
48. Tang, H., Wang, X., Xu, L., Ran, X., Li, X., Chen, L., Zhao, X., Deng, H.*, Liu, X., 2016. Establishment of local searching methods for orbitrap-based high throughput metabolomics analysis. Talanta 156-157, 163-71.
49. Yao, R., Ming, Z., Yan, L., Li, S., Wang, F., Ma, S.,Yu, C., Yang, M., Chen, L., Chen, L., Li, Y., Yan, C., Miao, D., Sun, Z., Yan, J., Sun, Y., Wang, L., Chu, J., Fan, S., He, W., Deng, H., Nan, F., Li, J., Rao, Z., Lou, Z., Xie, D., 2016. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469-73.
50. Wang, S., Lu, Y., Sun, X., Wu, D., Fu, B., Chen, Y., Deng, H., Chen, X., 2016. Identification of common and differential mechanisms of glomerulus and tubule senescence in 24-month-old rats by quantitative LC-MS/MS. Proteomics. doi: 10.1002/pmic.201600121.
51. Jiang, X., Feng, S., Chen, Y., Feng, Y., Deng, H.*, 2016. Proteomic analysis of mTOR inhibition-mediated phosphorylation changes in ribosomal proteins and eukaryotic translation initiation factors. Protein Cell 7, 533-7.
52. Huo, Y., Zheng, Z., Chen, Y., Wang, Q., Zhang, Z., Deng, H.*, 2016. Downregulation of vimentin expression increased drug resistance in ovarian cancer cells. Oncotarget doi: 10.18632/oncotarget.9970.
53. He, H., Liu, D., Lin, H., Jiang, S., Ying, Y., Chun, S., Deng, H., Zaia, J., Wen, R., Luo, Z., 2016. Phosphatidylethanolamine binding protein 4 (PEBP4) is a secreted protein and has multiple functions. Biochim Biophys Acta 1863, 1682-1689.
54. Gong, Y., Zhang, L., Li, J., Feng, S., Deng, H.,* Development of the Double Cyclic Peptide Ligand for Antibody Purification and Protein Detection. Bioconjug Chem 27, 1569-73.
55. Chu, Y., Zhu, Y., Chen, Y., Li, W., Zhang, Z., Liu, D.,Wang, T., Ma, J., Deng, H., Liu, Z. J., Ouyang, S., Huang, L., 2016. aKMT catalyzes extensive protein lysine methylation in the hyperthermophilic archaeon Sulfolobus islandicus but is dispensable for the growth of the organism. Mol Cell Proteomics. pii: mcp.M115.057778.
56. Jian, Y., Chen, Y., Geng, C., Liu, N.,Yang, G., Liu, J., Li, X., Deng, H.*, Chen, W., 2016. Target and resistance-related proteins of recombinant mutant human tumor necrosis factor-related apoptosis-inducing ligand on myeloma cell lines. Biomed Rep 4, 723-727.
57. Lu, Q., Liu, C., Liu, Y., Zhang, N., Deng, H., Zhang, Z., 2016. Serum markers of pre-eclampsia identified on proteomics. J Obstet Gynaecol Res.
58. Tang, H., Chen, Y., Liu, X., Wang, S., Lv, Y., Wu, D., Wang, Q., Luo, M., Deng, H.,* 2016. Downregulation of HSP60 disrupts mitochondrial proteostasis to promote tumorigenesis and progression in clear cell renal cell carcinoma. Oncotarget. doi: 10.18632/oncotarget.9615.
59. He, L., Nair, M. K., Chen, Y., Liu, X., Zhang, M., Hazlett, K. R., Deng, H.*, Zhang, J. R., 2016. The Protease Locus of Francisella tularensis LVS Is Required for Stress Tolerance and Infection in the Mammalian Host. Infect Immun 84, 1387-402.
60. Liu, Z., Hu, Y., Liang, H., Sun, Z., Feng, S., Deng, H.,* 2016. Silencing PRDX3 Inhibits Growth and Promotes Invasion and Extracellular Matrix Degradation in Hepatocellular Carcinoma Cells. J Proteome Res. 15, 1506-14.
61. Tang, H., Li, J., Liu, X., Wang, G., Luo, M., Deng, H. *, 2016. Down-regulation of HSP60 Suppresses the Proliferation of Glioblastoma Cells via the ROS/AMPK/mTOR Pathway. Sci. Rep. 6, 28388.
62. Zhao, D., Zhang, X., Guan, H., Xiong, X., Shi, X., Deng, H., Li, H., 2016. The BAH domain of BAHD1 is a histone H3K27me3 reader. Protein Cell 7, 222-226.
63. Wang, X., Tang, H., Chen, Y., Chi, B., Wang, S., Lv, Y., Wu, D., Ge, R., Deng, H.*, 2016. Overexpression of SIRT3 disrupts mitochondrial proteostasis and cell cycle progression. Protein Cell 7, 295-299.
64. Zheng, X., Ramani, A., Soni, K., Gottardo, M., Zheng, S., Ming Gooi, L., Li, W., Feng, S., Mariappan, A., Wason, A., Widlund, P., Pozniakovsky, A., Poser, I., Deng, H., Ou, G., Riparbelli, M., Giuliano, C., Hyman, A. A., Sattler, M., Gopalakrishnan, J., Li, H., 2016. Molecular basis for CPAP-tubulin interaction in controlling centriolar and ciliary length. Nat. Commun. 7, 11874.
65. Zhou, H., Li, J., Jian, Y., Chen, T., Deng, H., Zhang, J., Zeng, H., Shan, Z., Chen, W., 2016. Effects and mechanism of arsenic trioxide in combination with rmhTRAIL in multiple myeloma. Exp. Hematol. 44, 125-131 e11.
66. Song, P., Li, S., Wu, H., Gao, R., Rao, G., Wang, D., Chen, Z., Ma, B., Wang, H., Sui, N., Deng, H., Zhang, Z., Tang, T., Tan, Z., Han, Z., Lu, T., Zhu, Y., Chen, Q., 2016. Parkin promotes proteasomal degradation of p62: implication of selective vulnerability of neuronal cells in the pathogenesis of Parkinson's disease. Protein Cell 7, 114-129.
67. Lin, Z., Li, S., Feng, C., Yang, S., Wang, H., Ma, D., Zhang, J., Gou, M., Bu, D., Zhang, T., Kong, X., Wang, X., Sarig, O., Ren, Y., Dai, L., Liu, H., Zhang, J., Li, F., Hu, Y., Padalon-Brauch, G., Vodo, D., Zhou, F., Chen, T., Deng, H., Sprecher, E., Yang, Y., Tan, X., 2016. Stabilizing mutations of KLHL24 ubiquitin ligase cause loss of keratin 14 and human skin fragility. Nat. Genet. doi: 10.1038/ng.3701. [Epub ahead of print]
68. Jiang, X., Jiang, X.Y., Feng, Y., Wang, Q.T., Deng, H., 2016. Silencing of eIF5B Changes Proteostasis by Providing Negative Feedback on MAPK Signaling. PLoS One 11, e0168387.
69. Qu, H., Chen, Y.L., Deng, H., * Zhang, Z.Y., 2016. Identification and Validation of Differentially Expressed Proteins in Epithelial Ovarian Cancers Using Quantitative Proteomics, Oncotarget. doi: 10.18632/oncotarget.13077. [Epub ahead of print]
70. Ma, D.C., † Peng, S.G., † Xie, Z., * 2016. Integration and exchange of split dCas9 domains for transcriptional controls in mammalian cells. Nature Commun. 7, 13056.
71. Wang, T.T., Xie, Y., Tan, A., Li, S., Xie, Z., * 2016. Construction and characterization of synthetic microRNA cluster for multiplex RNA interference in mammalian cells, ACS Synthetic Biology 5, 1193–1200.
72. Cao, Z., Liu, C., Xu, J., You, L., Wang, C., Lou, W., Sun, B., Miao, Y., Liu, X., Wang, X., Zhang, T., Zhao, Y., 2016. Plasma microRNA panels to diagnose pancreatic cancer: Results from a multicenter study. Oncotarget. 7, 41575-41583.
73. Yuan, Ye., Ren, X.Y., Xie, Z., Wang, X., * 2016. A quantitative understanding of microRNA mediated competing endogenous RNA regulation. Quantitative Biology. 4, 47–57.
74. He, C., Li, G.P., Nadhir, D.M., Chen, Yang.,* Wang, X.,* Zhang, M.Q.,* 2016. Advances in computational ChIA-PET data analysis. Quantitative Biology. 4, 217–225.
75. 常海波,张 雪,张晓菲,张 翀,李和平*,邢新会*,2016.大气压冷等离子体与水溶液作用过程的数值模拟研究进展。化工进展,35,1929-1941.
国际学术交流:
1. Chen GQ, “Seawater Based Biotechnology”. Invited Lecture for ICBE 2016 (International Conference on Biomolecular Engineering: Biomolecular Engineering for Today’s Needs in Medicine, Materials and Sustainability). Singapore,January 5-7/2016
2. Chen GQ, “Synthetic Biology for Seawater Biotechnology”, Invited Lecture for SYNBIOCHEM, Manchester Inst of Biotechnology, Manchester University, Jan 10-13/2016
3. Chen GQ, “Seawater Based Industrial Biotechnology”. Keynote Lecture for The 7th International Symposium of Innovative BioProduction Kobe (iBioK), Kobe/Japan, January 25-28/2016
4. Chen GQ, “The PHAome”. Invited Lecture and Session Chair in the Joint POLY/PMSE Symposium on Sustainable Polymers, Processes, and Product Applications, 251st ACS National Meeting. San Diego/USA, March 13-17/2016
5. Chen GQ, “Seawater Based Industrial Biotechnology”. Keynote Lecture for Symposium “Molecular Biotechnology – from the Origins to New Horizons”, Graz, Austria, April 14- 15/2016
6. Chen GQ, “Synthetic Biology of Halomonas spp., Transforming the Chemical Industry to Biotechnology”. Invited by Paul S Freemont, Imperial College of London, June 6/2016
7. Chen GQ, “Synthetic Biology of Halomonas spp”. Invited Lecture and Co-Chair for Metabolic Engineering XI, Awaji Yumebutai Westin and International Conference Center, Japan, June 26-30/2016
8. Chen GQ, “Synthetic Biology of Seawater Based Biotechnology”. Invited by MIB Manchester University, Part of the EuroScience Open Forum (ESOF 2016). July 25-27/2016
9. Chen GQ, The “PHAome”. A Plenary Lecture to Remember Professor Bernard Witholt for his Great Contribution to the field. ISBP 2016, Marid/Spain, Sept 25-28/2016
10. Chen GQ, “Seawater Based Microbial Synthetic Biotechnology”. Invited Keynote Lecture (40 Minutes), 2016 AIChE Annual Meeting (Technical Section of Division 15C), San Fransisco, Nov 13-18/2016
11. Chen GQ, “Seawater Based Microbial Synthetic Biotechnology” (以海水为介质的微生物合成生物学). “Disruptive Technology in Bio-X Field” Sponsored by China Defense Science and Technology Information Center (CDSTIC). Beijing. Nov 23-25/2016
12. Chen GQ, “Halomonas spp as a New Chassis for Synthetic Biology”. Invited Lecture, Cold Spring Harbor Laboratory/Asia, Suzhou/China, Nov 28-Dec 2/2016
13. Yu Huimin, Jiao Song, Yang Ji, Zhangjing, Chen Jie, Shen Zhongyao. “Engineering nitrile hydratase by salt-bridges and disulfide-bridges.” 2016 annual meeting of Society of Biotechnology, Japan. Sep 28- 30, 2016. Toyama, Japan. Invited speaker.
14. Dai Junbiao, “Genome Engineering: From biological parts to whole genome synthesis” Invited Lecture, Cold Spring Harbor Laboratory/Asia, Suzhou/China, Nov 28-Dec 2/2016
15. Dai Junbiao, “Decode and reprogram the yeast genome”, Keynote Lecture, the International Conference on Metabolic Science (ICMS 2016), Shanghai, China, Oct 20-23/2016
16. Dai Junbiao, “Development of a “selector” system to identify strains with increased genetic variation after SCRaMbLEing”, Invited Lecture, the 13th International Symposium on the Genetics of Industrial Microorganisms, Wuhan, China, Oct 16-20/2016
17. Dai Junbiao, “Beyond Sc2.0: Re-engineer ChrXII ”, Invited Lecture, 5th Annual Sc2.0 and Synthetic Genomes Conference, Edinburgh, UK, July 8-9/2016
主办或协办国际学术会议:
1. “Synthetic Biology”. Chairman, Cold Spring Harbor Conference Asia (CSH) Dec 1-5/2016
2. “International Symposium on Biological Polyesters 2016 (ISBP 2016)”, Organizing Committee Member, Madrid, Spain, Sept 25-28/2016.
3. “Metabolic Engineering XI”. Co-Chairman, Awaji Westin and Conference Center on Awaji Island, Japan. June 26-30/2016
4. The 1st SynBioBeta China, June 14, 2016, Beijing China
5. 主办“10th Word Bioenergy Symposium”. Chairman, Dongguan (China) Dec 9-11/2016
国际影响:
Chen GQ
1. Associate Editor for “Microbial Cell Factories” (IF: 3.744) (Since September 2010)
2. Academic Editor for “Plos One” (IF 3.057) (Since July 2014)
3. Associate Editor for “Journal of Biotechnology” (Elsevier Publisher) (IF: 2.667) (Since Dec 2010)
4. Senior Editor for “Biotechnology Journal” (Wiley) (IF: 3.781) (Since January 2009)
5. Associate Editor for “Synthetic and Systems Biotechnology”. KeAi Publisher (Since Sept/2016)
6. Editorial Board for “Current Opinion in Biotechnology” (IF 8.314) (Since Nov 2010)
7. Editorial Board for “Biomaterials” (Elsevier Publisher) (IF: 8.387) (Since July 2007)
8. Editorial Board for “Biomacromolecules” (ACS) (IF 5.583) (Since January 2013)
9. Editorial Board for “Applied Microbiology and Biotechnology” (Springer-Verlag GmbH) (IF: 3.376) (Since May 2005)
10. Editorial Board for “ACS Synthetic Biology Journal” (ACS) (IF 6.076) (Since August 2011)
11. Editorial Board for “Metabolic Engineering” (Elsevier) (IF 8.201) (Since Jan 2016)
12. Editorial Board for “Microbial Biotechnology” (Wiley) (IF: 3.991) (Since February 2016)
13. Editorial Board for the “Artificial Cells, Nanotechnology and Biotechnology” (Taylor & Francis Inc) (IF: 2.024) (since 2008)
14. Editorial Board for “Synthetic Biology” (Oxford University Press) (Since April 5 2016)
15. Editorial Board for “Engineering Biology” (IET or Institute of Engineering and Technology, UK) (Since June 3/2016)
16. Executive Advisory Board for “Advanced Biosystems” (Wiley) (Since August 17 2016)
Liu DH
17. 2010年-至今 中国-巴西气候变化与能源技术创新研究中心主
18. 2015年-至今 中拉清洁能源与气候变化国际联合研究中心主任
19. 《Biofuels》编委
20. 《International Journal of Industrial Biotechnology》编委
Xie Zhen
1. Quantitative Biology杂志主编助理
2. 农业部转基因安全委员会委员
3. 中国人工智能学会生物信息学与人工生命专业委员会委员
4. 合成生物学青年学者论坛第二届组委会委员
Wang Xiaowo
中国生物工程学会首届主任委员
技术转化:
蓝水生物技术成立的“蓝晶生物”公司实现了第一次融资。
酶促非食用油脂制备生物柴油技术开发与集成放大,2016年中国石油和化学工业联合会技术发明一等奖
酶促生物柴油生产,2016年全球可再生能源领域最具投资价值的领先技术奖“蓝天奖”
校外经费:
重大专项:PHA作为可降解农膜(74万)
自然科学基金重大仪器专项:高通量微生物进化仪的研制(直接费用631.79万)
自然科学基金国际(地区)合作与交流项目:抗体-低分子量肝素偶联物治疗炎症性肠炎的创新药物设计及给药途径选择(直接费用166万元)
国家重点研发计划“七大作物育种”专项“”子课题(120万元)
自然科学基金面上项目:基于序列深度突变扫描高通量改造胞内色氨酸新生肽生物传感器响应特性(直接费用70万元)
国家重点研发计划“生物产业共性技术标准研究“专项“生物育种过程控制与检测技术标准研究”课题(290万元)